
MaNIS
The Mammal Networked Information System
Response to Reviewers’ Comments
We are pleased that the reviewers recognized the inherent value of this proposal to a broad constituency, from its potential to have substantial educational impact to its general utility to resource managers, policy makers, and research scientists. The acknowledgment that the proposed model is easily transferable to other disciplines and that cooperative georeferencing is a solution to a general problem underscores that this proposal will have broad impact beyond the community being immediately served.
In this resubmission we have emphasized design strategies to clarify the method of project implementation. We have also reworded portions of the proposal to clarify misunderstandings. A summary of our response to the four major concerns expressed by one or more reviewers is as follows:
Collaboration among seventeen collections in providing open access to standardized views of current, distributed data must be viewed in itself as an outstanding contribution from the museum community rather than a liability. Reviewers expressed concern that seventeen institutions could not be brought online within the three-year period of the proposal, particularly in the absence of a working prototype. This is an unfounded concern. A working prototype, the Distributed Database of North American Bird Data (Peterson et al., 1998), has been successfully implemented and tested for five collections over the last two years and that prototype forms the basis of this project. Two of the collections included in this proposal (KU and MVZ) are participants in that project. The seventeen institutions participating in this project will serve to demonstrate the viability of our enhancements to that network design (e.g., the addition of summary data for query optimization) for addressing issues of scalability in the existing prototype.
The addition of three months of salary for a second programmer, who will focus on server enhancements early in the project, not only addresses a reviewer’s concern that the proposal was too ambitious for a single programmer, but also the concern that visible results would not appear until the end of the project.
The availability of geospatial tools that permit analysis, modeling, and prediction of species distributions based on locality data and underlying environmental variables argues emphatically for the value of these data and the need to enhance current collections information by their inclusion. One reviewer’s concern that the cost of georeferencing was too high has been addressed in this resubmission by major changes to the georeferencing methods. First, the union of unique localities from all participating institutions will be assembled into a single resource accessible online to all participants. The lead programmer will automate the first pass of georeferencing of U.S. localities using SppFind (Vieglais, 1999), which will provide coordinates for roughly 70% of these localities. Using the online resource, participants will be able to verify and amend the automated coordinates at an estimated rate of 20 localities per hour. In-house experience in georeferencing at the Museum of Vertebrate Zoology, the California Academy of Sciences, and La Comision Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO) has provided consistent and reliable estimates of the rates at which the remaining localities can be georeferenced. These rates (9 per hr for the remaining U.S. localities, 6 per hr for non-U.S. North America, and 3 per hr for non-North America) are being applied consistently among the participating institutions in this resubmission. These changes to the methodols reduce the georeferencing effort to 48% of that of the original approach.
Finally, the ANSI/NISO Z39.50 standard for information retrieval has already proven successful in enabling distributed data sharing for natural history collections in the ornithological (Peterson et al., 1998) and ichthyological communities (FISHNET, 2000). It is not our intention to create a new protocol for the distribution of natural history data, but rather to provide enhancements to the existing, robust architecture that will facilitate data discovery, data availability, and network scalability.
Results of Prior Support
DEB-9630909: "Collections Information System Re-engineering, Museum of Vertebrate Zoology, University of California, Berkeley", plus REU supplements, August 15, 1996-August 31, 2000. PI Barbara R. Stein, Co-PI David B. Wake, $271,849.
Following a one-year effort in information modeling and requirements analysis, the Museum of Vertebrate Zoology (MVZ) obtained funds to implement a new collections information management system that would integrate specimen label data across taxonomic disciplines at several levels of conceptual and physical organization (Blum and Stein, 1995). The timing of that request coincided with the campus deactivation of the Museum’s 20-year-old database platform and the availability of more modern database system support. The strength and uniqueness of the MVZ data model was the integration of multiple specimen catalogs into a single processing environment and the ability to track individual specimen parts and other collection objects. Supplemental REU awards were made to address the issue of accessibility to the vast quantity of intimately-related data that are not contained on specimen labels and that did not yet exist in electronic form (e.g., field notebook pages and archival photographs). The replacement system has been designed to maximize the utility of information associated with the MVZ collections in support of research, biodiversity analyses, education, and management of collections. Accordingly, with support and assistance from the Berkeley Digital Library Project (DLP, 1999a), all MVZ specimen data and images of related archival materials may now be accessed electronically (http://elib.cs.berkeley.edu/mvz/). This project has revolutionized data access and our role as a data provider (see Impact of the Project, below). In addition, several institutions have incorporated the fruits of the re-engineering effort into their own systems (e.g., electronic taxonomic authority files and the MVZ data model (UAM, 1999; MVZ, 1998). This project represented conceptual and technical advances in the management of natural history collections’ information that has had a substantial impact in the community.
Project Goals
"The key to a cost-effective solution to the biodiversity crisis lies in the collaboration of museums, research institutions, and universities…" (Reaka-Kudla et al., 1997).
The crisis — The magnitude of human impacts on the ecological systems of Earth is overwhelmingly apparent. Human populations and their use of natural resources are increasing at an unsustainable rate. The urgent and unprecedented challenge is to understand biological systems in all their complexity in order to preserve them in a sustainable fashion.
The widespread distribution of valuable research specimens and authoritative information found in museum collections is frequently an impediment to systematic research as well as to understanding the role that many species play within the framework of global biodiversity. For example, ecological information from natural history observations is critical for developing policies to preserve threatened and endangered species, but attempts to extract this information are hindered by the lack of data integration and other obstacles to data discovery.
The solution — Harnessing
the accumulated knowledge of Earth’s biological diversity is fundamental
to sustaining global biological systems for the benefit of all of its inhabitants.
Research collections in museums and universities are the authoritative
source of knowledge about the identity, relationships, and properties of
species with which we share this planet. As such, collections play a central
and critical role in the conservation and sustainable use of biodiversity.
The potential contribution of specimen data to systematic, genomic, and
ecological analyses is orders of magnitude greater when information is
made easily accessible via distributed database systems that inter-operate
than when stand-alone systems are used (Blum, 2000). Imagine being able
to obtain data on millions of specimens of global geographic scope from
records housed across North America by submitting a single query on the
Internet. The traditional alternative has been to contact major collections
to discover the existence of relevant material and to request associated
data. This is a laborious and time-consuming task not only for the researcher,
but also for the curatorial staff providing the data. Even so, the researcher
is still faced with the challenge of integrating those data he or she is
lucky enough to obtain. Even after integration, most researchers with biogeographic
interests are faced with the enormous task of determining geospatial coordinates
for specimen localities prior to data analysis. In addition, important
specimen records housed in smaller or less well known collections are often
overlooked or neglected during this process.
|
Acronym |
|
Holdings |
georef |
to georef |
|
|
|
|
|
|
|
Access on WinNT |
|
|
|
|
|
|
Access on Win95 |
|
|
|
|
|
|
C/BASE 4.3 on DEC PRIORIS |
|
|
Natural History Museum |
|
|
|
Access, Specify on Win95 |
|
|
|
|
|
|
Access on Win95 |
|
|
Museum of Natural Science |
|
|
|
MacCurator, 4th Dimension on MacOS 8 |
|
|
Museum of Southwestern Biology |
|
|
|
Access on WinNT |
|
|
|
|
|
|
Argus, Sybase on WinNT |
|
|
Museum of Vertebrate Zoology |
|
|
|
Sybase on SunOS |
|
|
Slater Museum |
|
|
|
4th Dimension on MacOS 8 |
|
|
|
|
|
|
Access on Win95 |
|
|
|
|
|
|
FoxPro on Win95 |
|
|
|
|
|
|
Oracle on Solaris |
|
|
Museum of Zoology |
|
|
|
Filemaker Pro on Mac or Win |
|
|
|
|
|
|
MINARK 4.0 on DOS 5.0 |
|
|
|
|
|
|
Access on Win95 |
|
|
Burke Museum |
|
|
|
4th Dimension on MacOS 8 |
The proposed development of a distributed database of mammal specimen data represents a major collaborative effort and commitment by the participating institutions (see table above). The aggregation and integration of specimen data from their collections will facilitate access to information about Earth’s biological resources and will promote national and international scientific exchange of biodiversity information (Convention on Biological Diversity, 1992). The integration of these data across genomic, geospatial, and environmental domains will create new classes of biotic information for computational analysis and modeling (Krishtalka and Humphrey, 2000). Specifically, access to specimen data stored in natural history collections via the proposed network represents a significant step in the development of interoperable databases and will provide a foundation for future enhancements of bioinformatics capacities in the 21st century (PCAST Report, 1998; Pennisi, 2000). The importance of current and accurate specimen data for analysis by researchers and for conservation of biodiversity and ecosystems is paramount. The ability to provide those data easily, cost effectively, and globally through a distributed database as recommended by the Global Biodiversity Information Facility (Edwards, 1999; Edwards et al., 2000) is addressed by this proposal.
Scope and timeliness of the proposal — Seventeen North American museums will participate in this project. Together, these collections currently house more than 1,411,000 mammal specimens of global geographic scope, representing all orders and a majority of currently recognized families and genera. Other institutions, including those that have already expressed interest in joining the network, will be welcome to do so using their own resources once the network is functional and an installation package has been made available online.
The wealth of specimen data represented by this collaboration is of real and immediate value to the education, research, and conservation communities (see Impact of the Project, below). However, access to these data by other than the research community does not currently exist. Effective use of the vast store of collections-based information is hampered by inefficient access mechanisms. At the same time, institutional resources are being squandered because curators and collections managers must respond individually even to the simplest requests for data. Making a large number of specimen records easily accessible in a single schema via a distributed database will 1) increase use of collections at lower cost to institutions and the communities they serve, 2) increase contributions that specimens and their associated data make to solving the global biodiversity crisis, 3) encourage development of new applications for ecological analysis and synthesis, and 4) encourage educational use of specimen data, an activity that has no historical precedent.
The concept of combining data from multiple natural history collections is not new. An early example of uniform access to natural history data is Neodat II (1997). This system allows users to query data that have been extracted from multiple institutions and copied onto centralized servers. This system avoids query traffic on the databases of the participants, but it requires ongoing maintenance of the centralized servers and removes data from the immediate control of the participating organizations.
The development of the Z39.50 profile for distributed natural history collections data (ZBIG, 1998) was prototyped in the Distributed Database of North American Bird Data (Peterson et al., 1998), and provided a major advantage over centralized data warehousing by using a standard for distributed information retrieval that simultaneously accessed data directly from institutional databases. The ichthyological community has joined forces with marine biologists to exploit this technology through development of FISHNET (2000), a distributed information system that links together aquatic collections around the world and assemble environmental coverages suitable for predicting marine species distributions.
It is our intention in this project to elaborate on the implementation of the Z39.50 natural history collections profile described above by 1) expanding its specimen profile to include additional specimen data attributes, 2) optimizing data discovery and query mechanisms, 3) directing query traffic to data repositories on servers separate from the institutional databases, 4) providing scripts that migrate data based on rules under the direct control of participating collections, and 5) easily accommodating additional mammal collections in the future. Our design addresses both the growing demand for biodiversity information and the functional requirements of the institutions willing to provide that information through an open venue. Simultaneously, implementation of this project will provide self-maintaining, Internet-accessible dictionaries of taxonomic and geographic names that will be valuable tools for collections worldwide (see Incorporate dictionaries, below). The proposed network design can be adopted subsequently by other taxonomic disciplines with simple changes to the data model, and the existing networks that already use the underlying Z39.50 technology can benefit from the enhancements developed in this project.
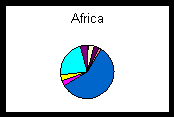
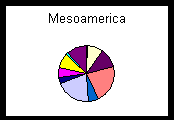
Well-documented, georeferenced collecting events are crucial to biogeographic data and biogeographic analyses clearly demonstrate the advantages of uniting information via a distributed network (Peterson et al., 1999). Geospatial mapping and modeling of collections data expose outliers in the database that either need correction if erroneous or warrant further study if valid. Georeferenced specimen localities from diverse collections increase the number and nature of questions to which biodiversity data can be successfully applied. The pie charts below show percentages of the total number of specimens from three geographic regions held by participating institutions. The pie chart of Africa represents a situation in which a large, well-known institution, in this case the Field Museum, holds a majority of specimens from a given region of the world. However, the localities represented by one museum for a given region may be incomplete or biased depending on the history of research expeditions that have been conducted. Collecting may have been confined to a local region, to a particular habitat type, or to a specific era. Data from several institutions will complement those from collections with large holdings by increasing not only the total number of specimens, but also the variety of collecting events. The pie chart of the combined holdings from Oceania illustrates that the contributions of a smaller collection, in this case the Bernice P. Bishop Museum, can be crucial to biogeographic investigations. The pie chart from Mesoamerica shows that specimens may be equally represented among a host of institutions, so that to neglect any one could result in a serious omission. The union of geographic data points from many collections ultimately will present a more complete and accurate picture of past and present species distributions. Sophisticated distribution prediction tools such as GARP (1997) demonstrate the power that combined data sets can bring to research and conservation issues (e.g., Peterson et al., 1999).



The Museum of Vertebrate Zoology is uniquely suited to undertake the development of the proposed Mammal Networked Information System (MaNIS) as it has been a leader among the participants testing the Distributed Database of North American Bird Data (Peterson et al., 1998). The Museum also has recruited the developer of that prototype (Dave Vieglais) to make the enhancements set forth in this proposal. In addition, having recently finished the re-engineering of the MVZ collections database management system, the lead programmer has an intimate understanding of the challenges involved in data modeling and data migration. By assisting the University of Alaska Museum in its migration to the MVZ data model, he has gained considerable insight into addressing these issues at other institutions.
The mammal community is equally suited for this project at the present time. The discipline encompasses a relatively stable taxonomy; the taxa are well-known, and they follow a generally-accepted taxonomic authority that has been rendered into electronic form (Wilson and Reeder, 1993), which is used as the basis of the taxonomic entries in the Integrated Taxonomic Information System (ITIS, 2000). In addition, among taxonomic disciplines, mammal collections have achieved a high degree of computerization of their specimen data (Hafner et al., 1997). Under the auspices of the American Society of Mammalogists, metadata documentation standards for data processing in mammalogy (ASM, 1996) were developed more than two decades ago (Williams et al., 1979) and have been implemented by most institutions that house mammal collections. Cooperation within the community, as evidenced by acceptance of these standards and by the large number of institutions interested in this project, indicates recognition of the value of the proposed project and bodes well for the future growth of the network.
Network Design Justification
The goal of this project is to create a network of distributed specimen collection databases that is 1) built on proven technologies, 2) scalable to include a large number of additional collections, 3) simple and low-cost to implement and maintain, 4) maximally accessible to users, and 5) easily adaptable to other disciplines. The details of the network design are elaborated in Project Implementation, below.
As described in Scope and timeliness of the proposal (see Project Goals, above), MaNIS uses the technologies developed and tested in the Distributed Database of North American Bird Data (Peterson et al., 1998). Implementation of the proposed network will provide and test enhancements to the existing technologies that overcome its recognized inefficiencies and scalability issues. The ANSI/NISO Z39.50 standard offers a stable solution for information retrieval with well-defined cross-domain search and retrieval, resource discovery, and error-handling mechanisms. Adoption of the requirements and standards of Z39.50 does not preclude the future migration to alternative technologies such as XML over HTTP. Instead, it provides a proven, robust framework with which to create an effective, functional resource now while preserving parity with other similar networks now in operation.
The network architecture must be simple, low-cost, and must conduct its operations with a minimum of hands-on maintenance if it is to endure without outside financial support and serve as a model for similar networks worldwide. When maintenance is required, it must be straightforward so that participating institutions can accomplish it themselves. Each participating institution’s hardware will consist of a single NT server and an Internet connection dedicated to this project. A dedicated server is essential in order to optimize the speed and efficiency with which data are returned to the user, and to avoid traffic and security risks to working institutional database systems ("master databases"). Each MaNIS server will store data in a no-license Microsoft® Database Engine (a "repository") on its dedicated server. The network architecture is able to accommodate other hardware and software configurations, but the uniformity of dedicated servers proposed for the participants in this project will minimize the programming effort required to bring collections online and will establish a user community with common technical issues. By using dedicated servers for this project the efficiency of data retrieval will be maximized and conflicts with existing working environments will be avoided. By designing for flexibility and implementing with efficiency, MaNIS will constitute a simple, robust, large-scale distributed network solution that is attractive to data providers, to users, and to other disciplines that have similar fundamental goals.
Specimen data from each participating institution will be migrated automatically on a weekly basis from each existing master database to a repository on a MaNIS server through scripts customized for that institution’s DBMS (e.g., Access, Sybase, Oracle), operating system (e.g., Unix, WinNT, MacOS), and institution-specific rules (e.g., do not make specific localities for endangered species publicly available). A free web server on each MaNIS workstation will hold a MaNIS network web page through which queries can be submitted to the network. A Z39.50 server on each MaNIS workstation will process queries from the web server, from other Z-clients, and from other MaNIS servers on the network. No additional hardware and software for backing up the MaNIS workstations will be needed. The configuration and data will be easily reproducible because 1) all software components will be easy to reinstall without customization from other MaNIS servers, 2) custom scripts for migrating data will reside on the computers that host the master databases, and 3) it will be possible to migrate data to a MaNIS repository from its master database on demand.
Project Implementation
Following is an overview of the steps necessary to build a large-scale, enhanced network based on the existing avian prototype (Peterson et al., 1998), which was designed by Dave Vieglais and which produced the Species Analyst (1998). Specific tasks within each step are detailed in the sections below.
The lead programmer will assemble copies of all relevant data from the databases of participating institutions to create dictionaries of unique higher geography and specific locality. These dictionaries will be Internet-accessible to facilitate cooperative georeferencing and to avoid redundancy of effort among collections. Georeferencing will begin immediately when this step is accomplished.
While georeferencing proceeds, the lead programmer will create scripts to migrate data from master databases to repositories for an initial group of institutions. Simultaneously, Vieglais will create enhancements to the existing Z39.50 server software. When these tasks are complete, a functional network of mammal collections’ data will have been created. The subsequent development of web interfaces and usage monitors for this enhanced network will make these data publicly available to the user community.
From this point on, project development will be an iterative process; scripts will be written to bring additional institutions online and the network will be tested and refined to optimize for the increased load. Migration scripts for these remaining institutions will require substantial programming effort given the arcane and disparate nature of many of the systems currently in use (see table above).
Finally, documentation and a suite of software in the form of an installation package will be created and made freely available over the Internet so that additional institutions can join the network. Other networks and disciplines will be encouraged to adopt and adapt the technologies developed in this project for their own use.
Implement specimen data model — Among the goals of MaNIS is to allow access to detailed specimen data in addition to the biogeographically-relevant data used in the current ANSI/NISO Z39.50 natural history profile (Vieglais, 1998). Access points (i.e., fields that can be queried) and elements (i.e., fields that are only returned in results) in the current profile are derived from the ASC information model (ASC, 1993), are independent of taxonomic discipline and include the following: Institution Code, Collection Code, Catalog Number, Lot Count, Kingdom, Phylum, Class, Order, Family, Genus, Species, Subspecies, Year, Month, Day, Collector, Specific Locality, Country, State/Province, County, Latitude, and Longitude. We propose to add elements for specimen parts, tissues, preparation method, karyology, type status, sex, age, collector’s number, stomach contents, habitats, lat/long source reference, lat/long maximum error distance, and geodetic datum to the data model and to document the format of these points. This documentation will serve as a guide to other disciplines wishing to extend the model. Given that data standards are already accepted within the mammal community (ASM, 1996), these additions will be straightforward to implement.
Coordinate georeferencing activities — Among data already in digital form, the number of records that are georeferenced is slowly increasing, but is still woefully incomplete. Only 21.7% of the total number of mammal specimens housed in the participating collections are currently georeferenced. Determination of latitude and longitude for specimen records will make these data valuable to a much broader segment of the research community (e.g., ecologists, phylogeographers, and conservationists). On a specimen level, this task would be daunting. However, by focusing on distinct localities rather than on individual specimens, the scale of the georeferencing problem can be made much more tractable. The creation at the start of this project of an online geographic dictionary (see Incorporate dictionaries, below) composed of the union of distinct geographic localities, including latitude and longitude, from the master databases of the participating institutions will reduce the scale of the georeferencing task. The MVZ and others have found that the number of distinct localities in their databases is roughly one-sixth to one-tenth of the total number of specimens in their mammal collections. Combining localities from all of the collections may reduce this ratio even further. An even more substantial reduction of effort is possible if locality coordinates can be applied programmatically and then verified rather than being determined one by one. SppFind (Vieglais, 1999) is one example of freely available software that can parse a locality string in search of a geographic named place from a digital gazetteer, in this case the Geographic Names Information System (USGS, 1997). Experience with the software shows that it can provide coordinates for 70% of typical specimen localities in the U.S.
After automating the first pass at georeferencing the resulting estimated coordinates will need to be verified. In anticipation of this need, the MVZ and the Berkeley Digital Library Project are currently collaborating to modify an existing tool that displays specimen occurrence data super-imposed on geospatial layers, including USGS topographic maps (DLP, 2000). The modifications to the tool will allow web-based queries on the locality dictionary to plot the results as circles centered on the coordinate and with a radius corresponding to the accuracy of the determination. The locality strings corresponding to the mapped occurrences will be visible by clicking on the plotted points. By automatically viewing points in association with the geographic named places from which they were estimated, users will be able to verify the plotted points quickly, make changes to the coordinates directly in the locality dictionary if necessary, and mark the record as verified. Experience suggests that localities can be verified using this method at a mean rate of 20 per hour.
Participants have recognized the value of collaborating in the task of georeferencing. Every institution faces similar problems in this realm and limited resources discourage duplication of effort. Although the primary goal of each institution will be to make sure that all of the localities from their own collections are georeferenced, the georeferencing workload will be shared between participants. Many participants have expressed their willingness to georeference all localities in geographic regions for which they have expertise. Furthermore, the nature of the online georeferencing tool will make it easy to verify coordinates for all of the localities that appear on a given topographic map, regardless of the institutional origin of those localities.
Localities that cannot be georeferenced automatically can be determined using a variety of other resources. With commercial software such as Terrain Navigatorä (MAPTECH, 1998), the MVZ has been able to georeference with metadata (e.g., accuracy, data source, datum) 99% of California localities at a mean rate of 9 localities per hour. Similar software is now freely available on the Internet (Topozone, 2000). Where USGS-related tools do not apply, slower rates of georeferencing have been determined through experience. Non-U.S. North American localities and non-North American localities can typically be georeferenced at rates of 6 and 3 per hour, respectively. Localities that will require significantly longer to georeference than these typical rates (ca. 1%) will be set aside as low priority, to be determined only if the allocated resources permit. Estimates of the number of equivalent weeks required to complete georeferencing for each participating collection are shown in the table above (see The Solution). These estimates are derived from the counts of non-georeferenced distinct localities in the three geographic categories mentioned above.
Write scripts to update specimen data in MaNIS repositories — As mentioned previously, identical MaNIS repositories will house the data that become publicly available through the network. These data will be derived from scripts that regularly migrate specimen data from the master databases to the standardized schema in the MaNIS repositories. The benefits of this approach are many, and include the following: 1) public access to specimen data will remain under the control of the individual institutions through customized scripts, 2) rules governing public access can differ among institutions, 3) rules can be changed by an institution at any time with no bearing on the function of the network, 4) public traffic will not affect master databases or increase network security risks for the participants, 5) the scheduling of updates is under the control of each institution, 6) master database upgrades, down-time, or re-engineering projects will not affect the public availability of data, 7) maintenance of the summary-level data can be optimized for query speed without affecting server loads or data concurrency, and 8) the software and network architecture used and developed in this project will not preclude the use of "live" views of data on working databases. The costs of achieving these many benefits include the initial investment of time to create the data migration scripts and the institutional commitments to maintain them. We are confident that the benefits to be gained by participating will more than compensate for the simple maintenance of individual servers.
Though there will be many migration issues in common among the participating institutions, the data migration scripts will be tailored for each institution due to the differences among collections' database schemas, database management systems, operating systems, and public access rules. Travel by the lead programmer to each of the institutions will be necessary to facilitate the implementation of the data migration scripts. As the project progresses, scripts written for one institution can be used as the basis for scripts at other institutions with similar systems. The data migration scripts that are developed will be incorporated in a knowledge base that can be used by future implementers (see Create installation package, below). The participating institutions are committed to working with the lead programmer and will provide expertise and technical assistance whenever possible. Letters of commitment to this project are on file in the MVZ.
Design summary data model — In addition to upgrading the ZBIG profile (Vieglais, 1998) so that the MaNIS servers can query on a greater number of fields and return more information, specimen data summaries and MaNIS server summaries will be added to the repositories in order to increase the speed with which end-users get results. The specimen summary tables will contain information such as counts of specimens by institution, geographic location, and taxonomy, whereas the MaNIS server summaries will contain information about the MaNIS servers themselves (e.g., institution, IP address, date and time of last data update, contact person, and email address). A preliminary query to the specimen summary on a local MaNIS repository would be able to answer the question, "Which institutions on the network have specimens of a given taxon from a given country?" Since the query is made from the same machine on which that information resides, the resulting list of institutions will be returned quickly. The full specimen query will then be automatically sent out only to those institutions on the list returned by the preliminary query. This will reduce network traffic and eliminate the need to wait for responses from institutions that do not have relevant specimen data. The benefit of this optimization will increase dramatically as the number of collections, the number of specimen records, and the number of users on the network increase.
Develop system summary software — The specimen data and MaNIS server summary tables need to accurately and automatically summarize the contents of all MaNIS repositories on the network. Unlike the specimen data migration scripts which reside on the systems hosting the master databases, the summary maintenance scripts will reside on, and be identical for, all MaNIS servers.
The automated maintenance of summary specimen information in the MaNIS repositories will be a multi-step process. First, a MaNIS server will be notified when the script that exports data from its master database is finished. The exported, tab-delimited specimen data files will then be transferred to the MaNIS server and each exported record will be compared with the records already on that server. If a difference is found, a new specimen record will be inserted and the old one, if it exists, will be tagged for deletion. After the specimen record comparisons are complete, the required changes to the summary data based on the changes to the specimen record will be computed and stored in a table of summary changes. Each record in the table of summary changes will be compared with the existing records. If a difference is noted, a new summary record will be inserted with a timestamp and an old one, if it exists, will be marked for deletion. Once the summary table regeneration is complete, the data bearing the current timestamp (the summary changes) will be exported to a document in XML-Data format (W3C, 1998). This document will be forwarded to each of the other MaNIS servers on the network using HTTP POST. On receipt of the summary changes, each MaNIS server will create a temporary record set from the XML document and perform a batch-update of its own summary tables. After the update has been forwarded and processed by all servers on the network, the records flagged for deletion in the specimen and summary data tables will be removed from the original repository and the MaNIS server will begin to operate with the updated records. This scheme minimizes both the amount of information that is sent between servers (differences rather than entire tables) and the duration of the discrepancy between remote and local summaries.
It is possible that a remote server will not immediately respond when POSTed the new summary data. The MaNIS server sending the data (the source) will continue to POST at increasing intervals until a response is received or until the next system update becomes available. If a MaNIS server becomes damaged or misses more than a single update, that server will indicate the time of last update in its response to the MaNIS source POST. It will then be the responsibility of the MaNIS source to generate a result set that contains the summary records modified since the indicated time and forward them back to the requesting MaNIS server using the same HTTP POST mechanism. Since the entire process is triggered by the initial specimen data updates, it is important to schedule this to occur at off-peak hours. The scheduling may be set manually or determined dynamically by examining an update history log. By including an identification key with transactions, and by refusing POSTs from unknown sources, the chance of malicious damage to the summary tables will be minimized.
Incorporate dictionaries — The addition of summary-level management to the MaNIS servers will generate interesting and valuable emergent properties. For example, the need to summarize and coordinate information among servers will be met by the automatic creation and management of replicated dictionaries of terms. These electronic dictionaries will act as current and comprehensive lists of useful vocabularies for data on the network and will be supersets of the automatically generated summary data. We propose to create both geographic and taxonomic dictionaries. The geographic dictionary, essential for optimizing biogeographic queries, will include related terms of higher-level geography (e.g., continent/ocean, country, state/province, county) and it will accommodate institutional variants such as country = "Burma" and country = "Myanmar", though it will not relate them to each other. The synonymization of related terms across institutions would require continuous management that is not within the purview of this project. Instead, users will be given the ability to browse these dictionaries within a hierarchical framework for terms of interest to them; for example, a list of countries in Asia would display both "Burma" and "Myanmar" so that users would know to query on both of these terms if they want to be assured of getting complete results for that country. Also, the dictionaries will allow any interested user to see how others are using taxonomic and geographic terms.
Given the dynamic nature of dictionaries, it will be useful to have a mechanism to delete entries that are no longer in use on the network. The most obvious use will be to remove a typographical error from the dictionary once it has been corrected and is no longer in use by any of the participating collections.
In addition to its role in optimizing geographic queries, the geographic dictionary, in conjunction with the viewer used to verify coordinates for localities, provides a powerful tool for shared georeferencing efforts (see Coordinate georeferencing activities, above). The same combination of viewer and geographic dictionary could also be used to make visual investigations of such aspects of the combined data as geographic distribution of collections by the various participating institutions.
In addition to its role in optimizing taxonomy-based queries, the taxonomic dictionary has the potential to act as the foundation for a dynamic Global Species Database (Edwards, 1999). A Global Species Database (GSD) is an index of all known species for a given taxon that is created by linking relevant databases worldwide. It contains a taxonomic checklist of all species within that taxon, provides synonymies and taxonomic opinions, can be updated and altered over time, and references significant alternative taxonomies and their synonymies. The discipline of mammalogy is fortunate in having a widely accepted taxonomic standard embodied in Mammal Species of the World (MSW) (Wilson and Reeder, 1993). We will include the taxonomic records of MSW as the core of the taxonomic dictionary for this project and will flag these records as having MSW as their source. As each new server is connected and, subsequently, as server summaries are updated, new taxa will be added to this dictionary automatically. Subspecific epithets and common names (missing from MSW) will build on this core and be an integral part of the dictionary. When a MaNIS server processes a summary update, a script will return a list of differences between that institution’s taxonomy and the list of taxa in the network dictionary. We anticipate that this reporting mechanism will be an extremely useful curatorial tool for checking data in the master databases. Since it will be possible for anyone with web access to browse the dictionaries, we anticipate that outdated taxonomy and misidentifications will be found and corrected quite readily.
It is not in the scope of this proposal, nor is it our intent, to assume the management of an online taxonomic authority. The dictionaries, as proposed, merely manage and show a summary of the content of repositories on the network. Nevertheless, the proposed model can accommodate any number of managed authorities on dedicated servers connected to the network and others will be free to build on this foundation.
Design Worldwide Web interfaces — The ultimate goal of this project is to facilitate access to distributed natural history data by creating both an efficient and effective venue to help providers of these data meet an ever-increasing demand for information. Accordingly, web interfaces to the data in the MaNIS repositories will be developed to enable users to query by institution, by taxonomy, by geography, by collector’s name, by year, or by any combination of these and other specimen attributes. Results will consist of a summary report listing the institutions queried, the timestamps of their last repository updates, and record counts for the given query by institution. Specimen records in list form with institutional acronyms at the beginning of each row will follow the summary report. Users will be allowed to query for just type specimens if they choose, or just tissue specimens, or specimen records by part and preservation mode.
Due to its ubiquity, familiarity, and low cost, the Worldwide Web is the appropriate medium for public access to MaNIS. All MaNIS servers will be equivalent; each of them will support a web server that will communicate with all repositories on the network using the ANSI/NISO Z39.50 protocol. The redundancy inherent in this approach assures that data will be available from the MaNIS network even if one or more MaNIS servers is disconnected or otherwise unavailable at any given time. From web sites on any of the MaNIS servers, users will be able to browse through collective data dictionaries, make specimen count queries, find contact information and links to collection web sites, view specimen data from the combined holdings of all of the networked collections and download them in text, GIS, and Excel formats. Institutions also will be able to view and download reports on the use of their online collections data (see Develop usage monitor, below). Those interested in creating a similar network or in understanding how the network functions will be able to download documentation and server software from any of the MaNIS web sites (see Create Installation Package, below).
Develop usage monitor — An important measure of the value of a natural history collection is the extent to which it is used (NSF guidelines, 1999), and collections have traditionally maintained statistics on specimen data access. The access to data can be measured continually and automatically both through the web server and the Z39.50 server. Using a combination of these two mechanisms, the lead programmer will incorporate into each MaNIS server a usage monitor similar in function to the one designed by the Digital Library Project at U.C. Berkeley (DLP, 1999b). Daily statistics available to participating institutions will include the origin, number, and nature of queries to the system, which data were downloaded, and the number and provenance of specimen records in each result set. Summaries of query statistics will be archived monthly and will be available for downloading from each MaNIS server.
Create installation package — In order to make MaNIS as easily accessible as possible to other mammal collections and to other taxonomic disciplines, the documentation and software developed for this project will be assembled and made freely available from each of the MaNIS servers at the beginning of the third year of the grant. During the third year, the lead programmer will test the installation package using the final group of participating institutions. It is our goal that any institution with the appropriate hardware will be able to download the installation package, install the software (all of it free of licensing), generate a repository, and create migration scripts. They would then be ready to register and thereby fully participate in the network.
We recommend that an institution wishing in the future to join an existing network send a request via email to the contact persons for the servers already on the network. Each contact will verify the merit of the request and add the information about the new server to the MaNIS repository. Thereafter, the new server will be included in queries originating from any MaNIS server to which the new server information has been added. Although it will be technically feasible to register a new server on an existing network by having it POST an XML record that includes its essential details (e.g., IP address, port, contact person) to the servers already on the network, such free access to join would leave the network vulnerable to malicious providers.
Timeline
Estimates for the time required to enhance the Z39.50 server are based on the second programmer’s experience in creating the original ZBIG server (ZBIG, 1998). The estimates of the time to accomplish the remaining activities are based on the experience of the lead programmer in re-engineering both the MVZ’s and University of Alaska Museum’s (UAM) database management systems. These efforts included data modeling, creation of specimen databases, data curation and migration, business rule analysis and implementation, creation of data dictionaries, georeferencing, and development of web interfaces for public data access.
The first goal of the project will be to facilitate georeferencing among the participating institutions so that they can begin using efficient techniques from the start. This part of the project will include the assembly of copies of the relevant data from the databases of the participating institutions, the creation of initial geographic and locality dictionaries from them, the automated assignment of coordinates to U.S. localities, and the deployment of an online tool to verify localities that have coordinates assigned to them. Three months after the receipt of funding, the geographic dictionary and georeferencing tool will be placed on the web and cooperative georeferencing will commence. The rates for georeferencing unique localities are detailed in Coordinate georeferencing activities (see above). Based on these rates, all of the georeferencing covered by this proposal can be completed within the three-year timeframe.
Once cooperative georeferencing has commenced, software development will proceed on two fronts for the next nine months. During that time, the lead programmer will create data migration scripts for five of the seventeen institutions while the second programmer develops the enhancements for data dictionaries, summary data, and Z-server communications. Thus, at the conclusion of the first year of the project, the MaNIS network will consist of five fully functional nodes ready for testing.
Early in the second year of the project, a student will develop web interfaces and usage monitors for the network. During the course of the second year, the lead programmer will create migration scripts for the next six institutions while testing and refining the enhancements to the network.
Early in the final year of the project, an installation package that includes server, repository, and network documentation will be created. This package will then be tested using the remaining six institutions. The lead programmer will also create the migration scripts for these six institutions while testing and refining enhancements to the network.
Deliverables
Throughout the Project Implementation section of this proposal are interspersed valuable properties of the network and its design. The four most important and far-reaching of these are mentioned here in summary.
The proposed development of a distributed database of North American mammal collections will impact not only the participating institutions, but also the research community, the informatics community, the education community, and society as a whole. Data combined via a distributed network will reveal geographic, ecological, and taxonomic knowledge gaps that need to be filled and will allow for prioritization and cost-effective planning of conservation and research efforts. Analysis of these combined data sets will also reveal patterns and processes of evolutionary and ecological phenomena that have not been apparent in analyses based on more limited data. In addition, the union of collections’ data via a distributed network is a prerequisite to efficiently integrate this vast storehouse of knowledge with genomic, biotic, and geospatial data and with the application of this information to complex biodiversity problems which are presently intractable. Finally, the implementation of the Z39.50 protocol for natural history collections has already served as the focus for the development of at least one analytical tool, Species Analyst (1998); it is anticipated that others will follow.
Members of the informatics community wishing to expand this network globally will have at their disposal the data standards, the installation and management software, and the documentation that are developed for this project. It is anticipated that the data summary features of MaNIS also will be applicable to other collections of Z39.50 systems, such as GEO, bibliographic, and GILs servers. These same enhancements will be of immediate benefit to the existing distributed natural history data networks (i.e., the avian network on which this project is based, and FISHNET).
The benefits that will accrue to society in general will be the standardized, easy access to quality specimen data, and the repatriation of those data to countries from which the specimens were collected. MaNIS will provide the first real opportunity that many countries outside of North America will have to access a wealth of current information about their own faunas because the number of specimens housed in collections on this continent is often far greater than the number housed in non-North American collections. Because of this opportunity, both the North American Biodiversity Information Network (NABIN), a tri-national project (Mexico, Canada, U.S.) of the Commission on Environmental Cooperation created as part of the North American Free Trade Agreement, and the National Biological Information Infrastructure (NBII), developed under the auspices of the U.S. Geological Survey, have strongly endorsed this proposal. Letters are on file in the MVZ.
The availability of large data sets via the proposed network will provide unprecedented opportunities for education in biodiversity conservation worldwide. There is increasing interest in large databases of natural history information as resources for inquiry-based learning at all educational levels. For example, The University of Michigan has recently submitted two successful proposals, one to NSF and one to IMLS (Institute of Museum and Library Services) that rely on its extensive use of natural history collections to address this need. The intrinsic appeal of natural history to people of all ages and backgrounds, in particular, the appeal of the large, charismatic mammalian megafauna, makes a networked database of such specific information an unusually potent tool for teaching about science and the scientific method. With MaNIS, teachers and educators wishing to engage the public will have an unparalleled tool at their disposal for accessing dynamic factual information quickly and easily. Equally vital and strategic will be the ongoing training of researchers and technicians who will expand this network and who will ensure the long-term viability of Earth's biodiversity.
Museums that already have public access to their specimen data have seen increased use of their collections and a dramatic decrease in staff time needed to fill information requests. For example, the Museum of Vertebrate Zoology web site fulfilled 41,937 specimen queries, representing 19,001,503 specimen records delivered, in the first year after the inception of its public data access. This is up from 95 requests representing 160,471 specimen records delivered manually by its staff in the preceding year. All project participants stand to serve a similarly broad segment of the community while allowing their staff to focus on ensuring currency and accuracy of their data. In addition, specimen data housed in smaller collections will gain visibility that was lacking previously. Each participant will become an equal partner and provider of biodiversity information at costs lower than they now incur. By placing smaller collections on par with those in larger institutions, a greater geographic representation of taxa will be achieved and previously perceived discontinuities in data will be reevaluated. Collaboration among institutions in this effort also will enhance collection management efforts. For example, it is increasingly common for a museum to hold a voucher specimen whose tissue is housed in another institution. The availability of easy access to specimen data through MaNIS will facilitate the cross-referencing of such information and ensure more complete and accurate data records. In addition, the taxonomic dictionary that will be built as part of this project could be used as the basis for a dynamic electronic version of Mammal Species of the World, with subspecies.
The cost of adding an institution to this network will consist of the investment in appropriate hardware, the investment in learning to migrate data to a network repository, and the commitment to maintain these components in order to realize the benefits of participation in the network. In addition, the network will establish a knowledgeable community of participants with common goals and experiences. Adapting this effort to another discipline will require the simple implementation of data standards for that discipline in the networked database schema. It is further expected that the partnership being created among institutions involved in this effort will be extended to previously unimagined collaborations among researchers in disparate disciplines (e.g., systematists, ecologists, molecular biologists, and environmental planners).
To further inform the research community about MaNIS, we propose to host a workshop at an annual meeting of the American Society of Mammalogists during the course of this project. The objectives of that workshop will be to demonstrate the capabilities of the network, to answer questions regarding its implementation, and to foster additional participation.
Finally, though this project will be implemented as a cooperative effort among mammal collections, it must be emphasized that the developments represented by this project can be applied freely in and between natural history disciplines that share the common need to have public access to taxonomically- and geographically-defined datasets.
Institutional Contributions towards this Project
Institutional commitments to participate in the development of a distributed mammal database must be viewed as a major contribution to this project in light of the autonomy under which museums have traditionally operated. Prior investments in committing specimen data to electronic form and in georeferencing locality information represent another substantial contribution by participating institutions, particularly among smaller collections in which much of this work has been accomplished without external support.
The lead programmer who will be hired to create this distributed network has extensive experience in all of the issues encompassed by this project, as well as a great interest in bringing this project to fruition. John Wieczorek has implemented the MVZ’s collection information system from its data migration phase to the debut of its public web interface (MVZ, 1999). He has also participated in the testing and deployment of the Z39.50 Biological Implementers Group server (ZBIG, 1998), which will serve as the basis for this development effort, and the Kansas KDI project which will benefit from the resources made available during the course of this project. The MVZ has already contributed fourteen months of programmer salary for Wieczorek during the completion of these tasks, a cost contribution equivalent to $82,341.
The Museum of Vertebrate Zoology, which houses the largest mammal collection among the participating institutions and has the smallest proportion of georeferenced localities (see table above), has also agreed to commit 24 months of half-time salary for a laboratory assistant to complete this critical task, a cost savings to the project of $22,209. Similarly, the Field Museum has agreed to contribute three months salary, over and above the funds requested, to facilitate georeferencing of its important collection, a contribution of $XXXX. Two other institutions housing large and important mammal collections, the University of Michigan and the Los Angeles County Museum of Natural History, have agreed to match 7% and 50%, respectively, of their indirect costs, for a total contribution of $XXXX to this project. In addition, several of the other institutions that will be participating in this project have in-house technical staff who will be able to work with Wieczorek to bring their data online, and all participating institutions have curatorial staff who will familiarize the programmer with their databases and institutional rules. No additional salary is requested for these persons and their time should be viewed as a major contribution to this project. Some institutions, including the Museum of Vertebrate Zoology, also will contribute a server and/or ethernet connection dedicated to this project as part of cost sharing ( see individual institutional budgets).
The Museum of Vertebrate Zoology is committed to providing 400 sq. ft. of office space for Wieczorek, a dual-CPU Pentium that will be used for development, a single-CPU Pentium that will serve for application development, ethernet connections for these two machines, phone, fax, and administrative support. The MVZ will also provide space, additional Pentiums, ethernet connections and all necessary software for the research assistant who will work on web interface design and the individuals who will be georeferencing specific localities. In addition, the MVZ will provide a web server on which all software and documentation for the project can be made publicly available. Server support and administration will continue to be provided at no cost to the Museum or to this project by the Instructional and Collections Computing Facility (Effie Dilworth, System Administrator), the Museum Informatics Project (Tom Duncan, Director), and the Digital Library Project (Robert Wilensky, Director) on the Berkeley campus.
The University of Kansas will provide 100 sq. ft. of office space for the second programmer, as well as computers, phone, fax, high-speed Ethernet connectivity, administrative support, and all software needed for Z-server development in this project. As creator of the original Z39.50 profile, Dave Vieglais brings an intimate familiarity of the software to this project and a detailed knowledge of exactly what is necessary to implement the enhancements described in this proposal. He has already devoted considerable time as a pro bono consultant to Wieczorek and Stein on this project, a contribution that should not be minimized.
Project coordination, planning for the ASM workshop, and
interactions between the participating institutions and the programmers
will continue to be directed and facilitated by the PI. Stein is well known
to all of the senior personnel in this project and has extensive experience
overseeing the planning, development, and implementation of the Museum
of Vertebrate Zoology's data model, its current database management system,
its public access data interface, and the coordination and development
of this proposal submission.
References Cited
ASC (Association of Systematics Collections). 1993. An Information Model for Biological Collections, Report of the Biological Collections Data Standards Workshop, August 18-24, 1992. (gopher://kaw.keil.ukans.edu:70/11/standards/asc)
ASM. 1996. Documentation Standards for Automatic Data Processing in Mammalogy, version 2.0. American Society of Mammalogists, Committee on Information Retrieval. (http://www.clpgh.org/cmnh/mammals/collections/docstandX.html)
Blum, S.D. 2000. Overview of Biodiversity Informatics. (http://www.calacademy.org/all_species/biodiv_informatics.html).
Blum, S.D. and B.R. Stein. 1995. A New Collections Information System Under Development at The Museum of Vertebrate Zoology. ASC Newsletter, 23:65,70.
Conference on Global Biodiversity. 1992. Rio de Janeiro. (http://www.iisd.ca/linkages/biodiv.html)
DLP (Digital Library Project, U.C. Berkeley). 1999a. (http://elib.cs.berkeley.edu/)
DLP (Digital Library Project, U.C. Berkeley). 1999b. Database Query Statistics. (http://elib.cs.berkeley.edu/webstats/)
DLP (Digital Library Project, U.C. Berkeley). 2000. GIS Viewer 3.0. (http://elib.cs.berkeley.edu/gis/)
Edwards, J.L. 1999. The Global Biodiversity Information Facility: an international network of interoperable biodiversity databases. ASC Newsletter, June/August, 27:6-7.
Edwards, J.L., M.A. Lane, and E.S. Nielsen. 2000. Interoperability of Biodiversity Databases: biodiversity information on every desktop. Science 289:2312-2314.
FISHNET. 2000. (The FISHNET Distributed Biodiversity Information System). (http://habanero.nhm.ukans.edu/fishnet/default.html)
GARP (Genetic Algorithm for Ruleset Production). 1997. (http://biodi.sdsc.edu/)
GBIF (Global Biodiversity Information Facility). 1999. Science, 285:22-23; The New York Times, July 27, p. D2. (http://www.york.biosis.org/gbif/index.htm)
Hafner, M.S., W.L. Gannon, J. Salazar-Bravo and S.T. Alvarez-Castañeda. 1997. Mammal Collections in the Western Hemisphere: a survey and directory of existing collections. American Society of Mammalogists. (http://asm.wku.edu/committees/systematic_collections/systematic_collections_committee_docs.html)
ITIS (Integrated Taxonomic Information System). 2000. (http://www.itis.usda.gov/)
KDI (Knowledge and Distributed Intelligence). 1998. Knowledge Networking of Biodiversity Information.
Krishtalka, L. and P.S. Humphrey. 2000. Can natural history museums capture the future? BioScience, 50:611-617.
MAPTECHä. 1998. Terrain Navigator: High Quality USGS Topographic Maps on CD-ROM. Version 2.04. (http://www.maptech.com/topo/index.html)
MVZ. 1998. Museum of Vertebrate Zoology (MVZ) Collections Information System Re-engineering Project. (http://shanana.berkeley.edu/mvz/cis/index.html)
MVZ. 1999. Museum of Vertebrate Zoology Data Access. (http://elib.cs.berkeley.edu/mvz/)
Neodat II. 1997. The Inter-Institutional Database of Fish Biodiversity in the Neotropics. (http://neodat.org/)
NSF guidelines. 1999. Biological Research Collections Program Announcement. NSF 98-126. (http://www.nsf.gov/pubs/1998/nsf98126/nsf98126.htm)
PCAST Report. 1998. Teaming with Life: Investing in Science to Understand and Use America’s Living Capital. PCAST Panel on Biodiversity and Ecosystems. (http://www.whitehouse.gov/WH/EOP/OSTP/Environment/html/teamingcover.html)
Pennisi, E. 2000. Taxonomic Revival. Science, 289:2306-2308.
Peterson, A.T., J. Soberón and V. Sánchez-Cordero. 1999. Conservatism of Ecological Niches in Evolutionary Time. Science, 285:1265-1267.
Peterson, A.T., D.A. Vieglais, D.R.B. Stockwell, and A.G. Navarro. 1998. Distributed Database of North American Bird Data. National Science Foundation-funded project resulting in The Species Analyst.
Reaka-Kudla, M.L., D.E. Wilson and E.O. Wilson, eds. 1997. Biodiversity II: understanding and protecting our natural resources. Joseph Henry Press, Washington, D.C.
Species Analyst. 1998. (http://habanero.nhm.ukans.edu/)
(http://apps.internet2.edu/sept98/species.htm)
(http://habanero.nhm.ukans.edu/documentation/applications/SpeciesAnalyst/)
The TopoZone. Maps ala carte, Inc. 2000. (http://www.topozone.com/)
UAM (University of Alaska Museum). 1999. The Alaska Frozen Tissue Collection, a resource for investigations of northern mammals (DBI-9876837).
USGS. 1997. Geographic Names Information System. (http://nsdi.usgs.gov/products/gnis.html)
Vieglais, D.A. 1998. Prototype Z39.50 Interface to Biological Collections Databases (DEB 9808739). (http://habanero.nhm.ukans.edu/documentation/applications/CollectionAttributes.htm)
Vieglais, D.A. 1999. SppFIND. Freeware specific locality georeferencing software for U.S. localities contained in the USGS GNIS. (http://habanero.nhm.ukans.edu/Z.X/documents/findme)
W3C. 1998. XML-data. (http://www.w3.org/TR/1998/NOTE-XML-data/).
Williams, S.L., M.J. Smolen and A.A. Brigida. 1979. Documentation standards for automatic data processing in mammalogy. The Museum of Texas Tech University, Lubbock, TX.
Wilson, D.E. and D.M. Reeder. 1993. Mammal Species of the World: a taxonomic and geographic reference. Association of Systematics Collections and the Smithsonian Institution Press, Washington, D.C. (http://www.nmnh.si.edu/msw/)
ZBIG. 1998. An Experimental Z39.50 Information Retrieval
Protocol Test Bed for Biological Collection and Taxonomic Data. (http://habanero.nhm.ukans.edu/ZBIG/proposal.htm)
| John Wieczorek, 27 June 2001 |
Rev. 5 Jul 2005, JRW
|